2. 阜阳市疾病预防控制中心检验科, 安徽 阜阳 236030;
3. 马鞍山市疾病预防控制中心检验科, 安徽 马鞍山 243011;
4. 蚌埠市疾病预防控制中心检验科, 安徽 蚌埠 233000
 唑63.6%、23.5%、46.8%,氨苄西林/舒巴坦28.0%、9.2%、29.8%,庆大霉素34.7%、7.1%、34.0%,氯霉素22.9%、5.1%、21.3%,环丙沙星18.6%、4.1%、14.9%;2株EAEC和1株ETEC对亚胺培南耐药。55株EAEC分为35个ST型,优势基因型是ST10(14.5%);55株ETEC分为14个ST型,优势基因型是ST48(32.7%)和ST218(16.4%);15株EPEC分为12个ST型,优势基因型是ST28(13.3%)、ST517(13.3%)和ST2088(13.3%)。微进化树分析结果显示:55株ETEC中,马鞍山市分别有5、3、2、2株菌100%同源,合肥市有5株菌100%同源;55株EAEC中,阜阳市、滁州市各有2株菌100%同源;15株EPEC中,有2组2株菌100%同源,分离自马鞍山市。50株EAEC PFGE同源性聚类分析结果为:100%同源有2株,分离自蚌埠市,其余菌株同源性均在90%以下;21株EPEC的基因同源性均在75%以下;30株ETEC、3株肠侵袭性大肠埃希菌(EIEC)和5株ETEC+aEAEC的基因同源性达97.6%、93.6%各2株,其余菌株同源性都在90%以下。结论 本研究发现安徽省临床分离的DEC耐药情况较严重,其中EAEC的耐药率高于EPEC和ETEC,ETEC的耐药率在各致病型中最低。分子分型检测发现本地区临床分离的DEC基因型较分散,在各致病型中,ETEC的基因型相对集中。微进化树分析提示ETEC存在地区同源性感染。在分子分型技术上,MLST能区分出优势基因型和聚集性,优于PFGE分型。
唑63.6%、23.5%、46.8%,氨苄西林/舒巴坦28.0%、9.2%、29.8%,庆大霉素34.7%、7.1%、34.0%,氯霉素22.9%、5.1%、21.3%,环丙沙星18.6%、4.1%、14.9%;2株EAEC和1株ETEC对亚胺培南耐药。55株EAEC分为35个ST型,优势基因型是ST10(14.5%);55株ETEC分为14个ST型,优势基因型是ST48(32.7%)和ST218(16.4%);15株EPEC分为12个ST型,优势基因型是ST28(13.3%)、ST517(13.3%)和ST2088(13.3%)。微进化树分析结果显示:55株ETEC中,马鞍山市分别有5、3、2、2株菌100%同源,合肥市有5株菌100%同源;55株EAEC中,阜阳市、滁州市各有2株菌100%同源;15株EPEC中,有2组2株菌100%同源,分离自马鞍山市。50株EAEC PFGE同源性聚类分析结果为:100%同源有2株,分离自蚌埠市,其余菌株同源性均在90%以下;21株EPEC的基因同源性均在75%以下;30株ETEC、3株肠侵袭性大肠埃希菌(EIEC)和5株ETEC+aEAEC的基因同源性达97.6%、93.6%各2株,其余菌株同源性都在90%以下。结论 本研究发现安徽省临床分离的DEC耐药情况较严重,其中EAEC的耐药率高于EPEC和ETEC,ETEC的耐药率在各致病型中最低。分子分型检测发现本地区临床分离的DEC基因型较分散,在各致病型中,ETEC的基因型相对集中。微进化树分析提示ETEC存在地区同源性感染。在分子分型技术上,MLST能区分出优势基因型和聚集性,优于PFGE分型。2. Department of Laboratory Medicine, Fuyang Center for Disease Control and Prevention, Fuyang 236030, China;
3. Department of Laboratory Medicine, Ma'an- shan Center for Disease Control and Prevention, Ma'anshan 243011, China;
4. Department of Laboratory Medicine, Bengbu Center for Disease Control and Prevention, Bengbu 233000, China
致泻性大肠埃希菌(diarrheagenic Escherichia coli,DEC)仍是当前发展中国家引起急性胃肠炎的主要病原体之一,主要通过污染的食品或饮用水进行传播。可分为6种病理类型:肠致病性大肠埃希菌(EPEC)、肠侵袭性大肠埃希菌(EIEC)、肠聚集性大肠埃希菌(EAEC)、弥散黏附性大肠埃希菌(DAEC)、肠出血大肠埃希菌(EHEC)和肠毒素性大肠埃希菌(ETEC)。除DAEC无特异的毒力基因鉴定外,其余5种病理类型都可通过特异性毒力基因进行鉴定分型[1-2]。
DEC在夏秋季节常引起一些散发病例,偶尔也会发生一些聚集性病例。基因溯源在流行病学和疾病预防控制中非常重要,其中多位点序列分型(multilocus sequence typing,MLST)和脉冲场凝胶电泳(pulsed-field gel electrophoresis,PFGE)分型是较常用的基因溯源方法。PFGE分型选用适当的限制性内切酶在胶块中对整个细菌染色体进行酶切,360°电泳分离,最后根据分离的条带数和大小进行基因溯源[3]。MLST是对保守基因测序分型,分辨率高,数据重复性好,常用于细菌基因分型和进化研究[2, 4-6]。
本研究主要对安徽省连续2年临床分离的DEC分别用PFGE分型和MLST进行基因同源性分析,了解该地区DEC各致病型主要流行的基因型及同源性,并比对两种分型方法的优缺点。
1 材料与方法 1.1 材料 1.1.1 菌株来源2018—2019年安徽省9个地市哨点医院采集的腹泻患者粪便标本分离的DEC。
1.1.2 主要仪器和试剂聚合酶链反应(PCR)仪(PTC-200) 购自美国MJ公司,凝胶成像仪Gel-DocXR购自美国Bio-Rad公司,CHEF-Mapper型脉冲场凝胶电泳仪购自美国Bio-Rad公司。GoTaq Green Master Mix (Lot: M7122) 购于Promega公司,XbaⅠ限制性内切酶购于TaKaRa公司。
1.2 方法 1.2.1 药敏试验采用微量肉汤稀释法进行药敏试验,试验方法及药物的选择参照美国临床实验室标准化协会(CLSI)—2019[7]标准。挑取新鲜纯菌落3~5个,用无菌水配制成0.5麦氏单位的菌悬液,吸取60 μL加入12 mL肉汤管中充分混匀。向96孔药敏板中每孔加入100 μL(约5×105 CFU/mL) 的肉汤菌悬液,置湿盒中36 ℃培养20 h[8]。质控菌株:大肠埃希菌ATCC 25922和金黄色葡萄球菌ATCC 25923。
1.2.2 MLST随机对2018年临床分离的DEC进行基因测序分型。根据E.coli数据库提供的大肠埃希菌MLST方案,参考数据库和文献合成引物[8],配制50 μL PCR扩增体系:GoTaq Green Master Mix 2 μL,10 μmol/L的上下游引物各2 μL,无RNA酶水19 μL,DNA模板2 μL。PCR扩增条件参照文献[9]。收集PCR产物,纯化后进行测序[2]。将基因序列导入E.coli MLST数据库中获得相应的ST型,将7对管家基因序列按(adk-fumC-gyrB-icd-mdh-purA-recA)顺序连接后,用MEGA v6.0构建微进化树[minimum-evolution (ME) tree][4, 10]。
1.2.3 PFGE分型对2019年临床分离的DEC进行PFGE分型。①首先制备1% SeaKem Gold plug:调制4.0~4.5麦氏单位的菌悬液,吸取400 μL菌悬液至EP管中,每管中加入400 μL SeaKem Gold,充分混匀后加入plug模具中,室温凝固30 min;细菌裂解,将制备好的1% SeaKem Gold plug加入CLB裂解液中54℃温育2 h;②Plug清洗:用无菌超纯水洗2次,TE缓冲液洗4次;③DNA酶切:取出plug,切成约2.0 mm宽的小胶条,加入200 μL XbaⅠ酶切液,37℃水浴2 h;④酶切片段电泳:将酶切后的小胶条粘贴在梳齿上,倒入1% SKG,待胶块凝固后电泳[3]。Bionumerics 6.6软件分析。
2 结果 2.1 检出菌株情况2018—2019年,全省9个地市哨点医院共采集1 920例腹泻患者粪便,分离DEC 268株。病理类型为:EAEC 118株(44.0%),其中非典型EAEC(aEAEC) 80株(29.9%),典型EAEC(tEAEC)38株(14.2%);ETEC 98株(36.6%),其中ETEC 92株(34.3%),ETEC+aEAEC 6株(2.2%);EPEC 47株(17.5%),其中非典型EPEC(aEPEC)45株(16.8%),典型EPEC(tEPEC)2株(0.7%);EIEC 5株(1.9%)。病原菌来源地分布:马鞍山市78株(29.1%),阜阳市62株(23.1%),合肥市50株(18.7%),蚌埠市27株(10.1%),淮南市18株(6.7%),亳州市16株(6.0%),池州市7株(2.6%),滁州市6株(2.2%),芜湖市4株(1.5%)。
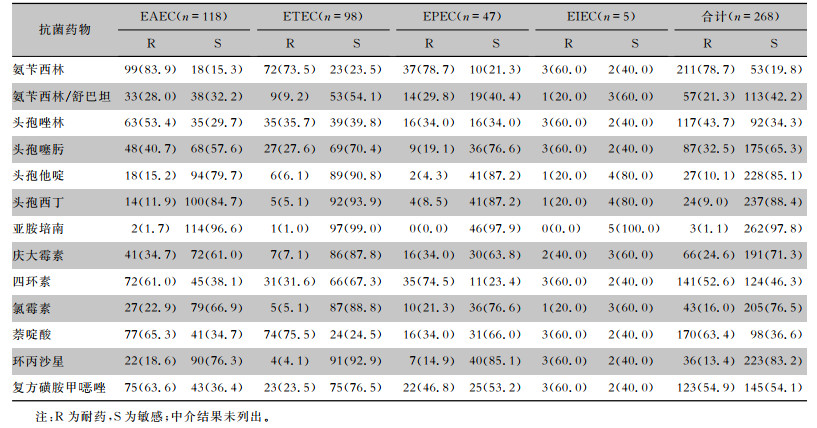
2.2 药敏结果268株DEC中,57.1%的菌株为多重耐药菌,对氨苄西林、氨苄西林/舒巴坦、头孢唑林、庆大霉素、四环素、氯霉素、萘啶酸、环丙沙星和复方磺胺甲
| 表 1 四种致病型DEC对13种抗菌药物的药敏结果[株(%)] Table 1 Antimicrobial susceptibility testing results of 4 types of pathogenic DEC to 13 kinds of antimicrobial agents (No. of isolates[%]) |
|
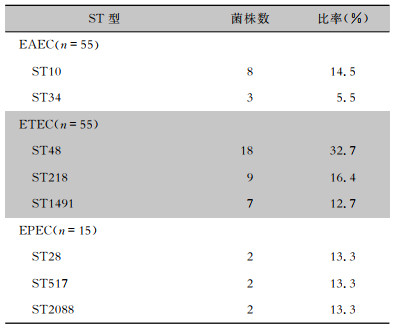
随机对2018年临床分离的127株DEC进行MLST,共获得60个ST型,其中EAEC和ETEC少数菌株存在ST48和ST394型交叉。55株EAEC分为35个ST型,优势基因型是ST10(14.5%);55株ETEC分为14个ST型,优势基因型是ST48(32.7%)和ST218 (16.4%);15株EPEC分为12个ST型,优势基因型是ST28、ST517和ST2088,各占13.3%;2株EIEC分为ST6和ST99。见表 2。15株EPEC微进化树分析有2组100%同源,其中1组4株(2株分离自马鞍山市,1株分离自合肥市,1株分离自芜湖市),另1组2株菌分离自马鞍山市,见图 1。55株ETEC微进化树分析有5组100%同源,其中1组5株菌分离自马鞍山市,另有2组2株菌均分离自马鞍山市,还有1组9株菌(5株分离自合肥市,3株分离自马鞍山,1株分离自阜阳市),见图 2。55株EAEC微进化树分析有4组100%同源,其中1组3株(2株分离自阜阳市,1株分离自合肥市),1组2株分离自滁州市,其余2组2株菌均分离自不同地方,见图 3。
| 表 2 主要致病型DEC的MLST结果 Table 2 MLST results of main pathogenic DEC |
|
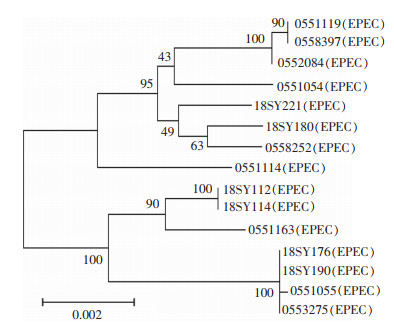 |
| 图 1 15株EPEC微进化树图(MEGA v6.0) Figure 1 Minimum-evolution tree of 15 EPEC strains (MEGA v6.0) |
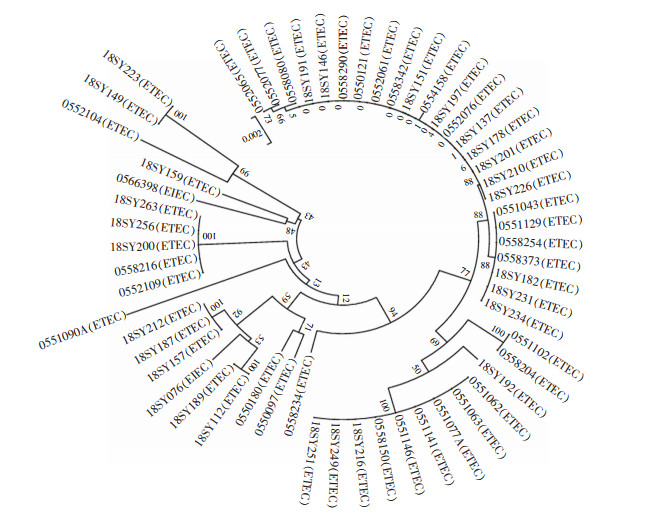 |
| 图 2 55株ETEC和2株EIEC微进化树图(MEGA v6.0) Figure 2 Minimum-evolution tree of 55 ETEC strains and 2 EIEC strains (MEGA v6.0) |
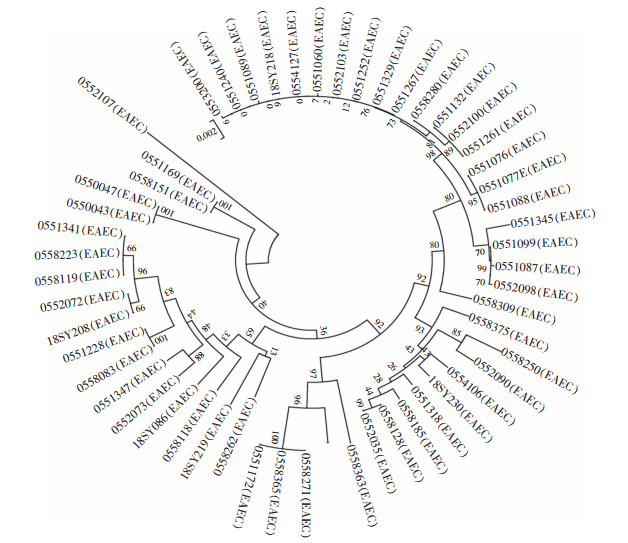 |
| 图 3 55株EAEC微进化树图(MEGA v6.0) Figure 3 Minimum-evolution tree of 55 EAEC strains (MEGA v6.0) |
2019年临床分离的109株DEC基因同源性聚类分析结果显示,50株EAEC(tEAEC 18株,aEAEC 32株)中有2株100%同源,分离自蚌埠市,其余菌株同源性均在90%以下,见图 4;21株EPEC同源性均在75%以下,见图 5。30株ETEC、3株EIEC和5株ETEC+aEAEC中,同源性达97.6%、93.6%各2株,其余菌株同源性均在90%以下,见图 6。
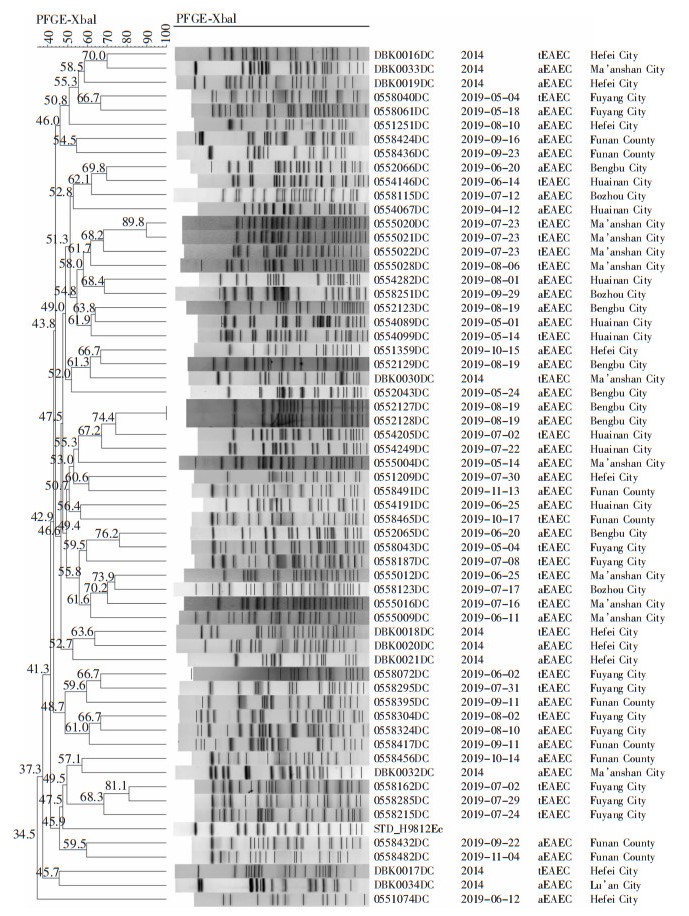 |
| 注:2019年安徽地区分离的50株EAEC与数据库中2014年已上传的10株EAEC聚类分析;Fuyang City为阜阳市,Funan County为阜南县,Hefei City为合肥市,Bengbu City为蚌埠市,Ma’anshan City为马鞍山市,Huainan City为淮南市,Bozhou City为亳州市,Lu’an City为六安市。 图 4 60株EAEC的PFGE同源性聚类图 Figure 4 PFGE homology clustering diagram of 60 EAEC strains |
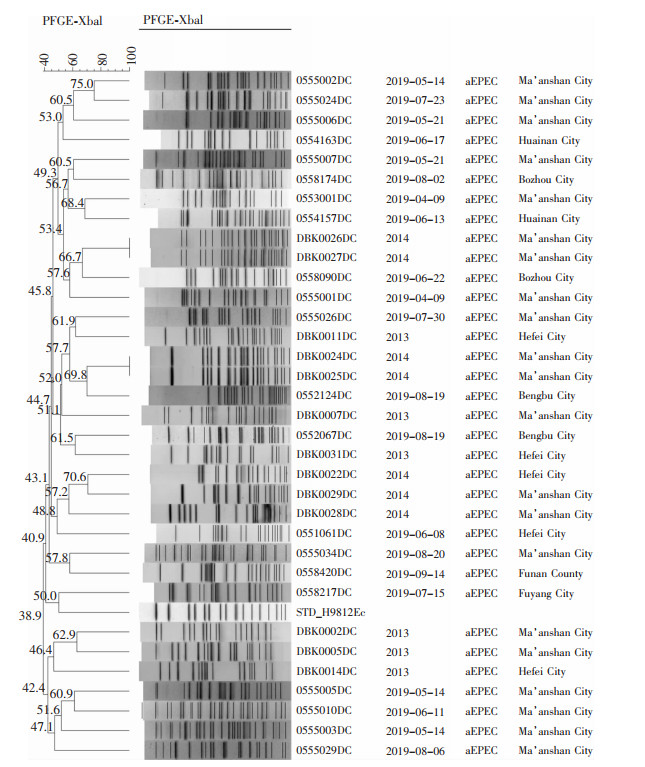 |
| 注:2019年安徽地区分离的21株EPEC与数据库中2013—2014年已上传的13株EPEC聚类分析;Fuyang City为阜阳市,Funan County为阜南县,Hefei City为合肥市,Bengbu City为蚌埠市,Ma’anshan City为马鞍山市,Huainan City为淮南市,Bozhou City为亳州市。 图 5 34株EPEC的PFGE同源性聚类图 Figure 5 PFGE homology clustering diagram of 34 EPEC strains |
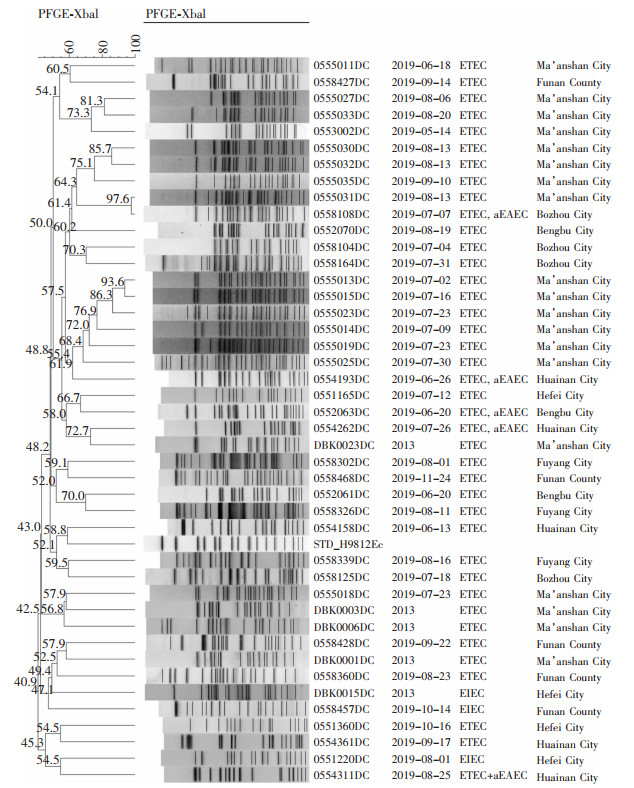 |
| 注:2019年安徽地区分离的30株ETEC、3株EIEC、5株ETEC+aEAEC与数据库中2013年上传的4株ETEC和1株EIEC聚类分析;Fuyang City为阜阳市,Funan County为阜南县,Hefei City为合肥市,Bengbu City为蚌埠市,Ma’anshan City为马鞍山市,Huainan City为淮南市,Bozhou City为亳州市。 图 6 43株ETEC和EIEC的PFGE同源性聚类图 Figure 6 PFGE homology clustering diagram of 43 ETEC and EIEC strains |
本研究资料显示,安徽地区流行的DEC存在4种优势毒株,其中以EAEC为主,占44.0%,其次是ETEC(36.6%)、EPEC(17.5%)和EIEC(1.9%),未从患者粪便中检测出EHEC,提示本地区流行的DEC主要是EAEC、ETEC和EPEC,与张子科等[11]统计的结果基本相吻合。
药敏试验结果显示,临床分离的DEC毒株多重耐药菌检出率高达57.1%,对氨苄西林、萘啶酸和头孢唑林的耐药率较高,耐药情况较严重,较之前的药敏监测结果略有升高[12],与相关研究[13-14]报道的结果近似,提示我国多省份DEC的耐药情况较严重。本研究发现EAEC对头孢唑林和头孢他啶的耐药率分别为53.4%、15.2%,明显高于ETEC的35.7%、6.1%,以及EPEC的34.0%、4.3%。EPEC对萘啶酸和头孢噻肟的耐药率分别为34.0%、19.1%,明显低于EAEC的65.3%、40.7%,以及ETEC的75.5%、27.6%。ETEC对四环素、复方磺胺甲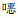
2019年复旦大学附属华山医院的监测数据[15]显示,耐碳青霉烯类肠杆菌目细菌(CRE)检出率为20.4%,肺炎克雷伯菌中耐碳青霉烯类菌株检出率为42.9%,临床耐药菌株尤其是CRE呈增长趋势。但DEC中近年才检出CRE,本研究在2013—2018年的监测数据中均未发现耐碳青霉烯类DEC,仅2019年发现2株EAEC和1株ETEC对亚胺培南耐药,后续将对其耐药机制和传播机制进行研究。
MLST结果显示,127株DEC共检出60个ST型,呈高度多态性。其中55株EAEC分为35个ST型,以ST10(14.5%)和ST34(5.5%)略占优势,与Okeke等[16]报道Nigeria临床分离的126株EAEC多达96种ST型的多态性分布相近。微进化树分析显示,有少数菌株呈100%同源,但空间分布广泛,提示无聚集性感染。50株EAEC的PFGE同源性聚类分析仅有2株出现100%同源性,分离自蚌埠市,其余菌株的同源性均小于90%。同样提示EAEC高度多态性分布,无聚集性感染。Li等[5]报道深圳地区分离的ETEC基因型较分散,ST218为相对优势基因型,本研究中55株ETEC分为14个ST型,优势基因型是ST48(32.7%)、ST218 (16.4%)和ST1491(12.7%),说明本地区临床分离的ETEC基因型较集中,ST48为主要优势基因型,提示不同地区ETEC基因型存在差异性。55株ETEC微进化树分析发现有多达5组100%同源,其中有多株菌分离自马鞍市,提示存在地区聚集性感染的可能性,但流行病学调查未发现暴发或聚集性病例。研究发现同源性感染存在时间差异,因此未出现聚集性病例,可能与菌株的克隆传播有关。30株ETEC PFGE同源性聚类分析发现,同源性达97.6%、93.6%各2株,其余菌株同源性在90%以下。15株EPEC分为12个ST型,呈多态性分布,其中以ST28(13.3%)、ST517(13.3%)和ST2088(13.3%)略占优势。15株EPEC微进化树分析显示,少数菌株呈100%同源性,空间分布广泛,提示无聚集性病例出现。21株EPEC PFGE同源性聚类分析显示同源性均在75%以下。
PFGE和MLST同为两种分子分型方法,两者各有优缺点,可以互为补充用于致病菌的分型和基因溯源分析。从本研究分型结果看,两者作用相同,均可用于分析致病菌的同源性,并且两种分型方法获得的结果也近似。但MLST是通过基因测序,其准确度要高于PFGE法,在本研究中MLST法对ETEC同源性的判断要比PFGE法更精准。但PFGE法的优势是普及程度高,可用于大范围多样本的分析判断。
利益冲突:所有作者均声明不存在利益冲突。
| [1] |
Croxen MA, Law RJ, Scholz R, et al. Recent advances in understanding enteric pathogenic Escherichia coli[J]. Clin Microbiol Rev, 2013, 26(4): 822-880. |
| [2] |
Yu F, Chen X, Zheng SF, et al. Prevalence and genetic diversity of human diarrheagenic Escherichia coli isolates by multilocus sequence typing[J]. Int J Infect Dis, 2018, 67: 7-13. DOI:10.1016/j.ijid.2017.11.025 |
| [3] |
Zhang SX, Zhou YM, Tian LG, et al. Antibiotic resistance and molecular characterization of diarrheagenic Escherichia coli and non-typhoidal Salmonella strains isolated from infections in Southwest China[J]. 2018, 7(1): 53.
|
| [4] |
Bagus Wasito E, Shigemura K, Osawa KY, et al. Antibiotic susceptibilities and genetic characteristics of extended-spectrum beta-lactamase-producing Escherichia coli isolates from stools of pediatric diarrhea patients in Surabaya, Indonesia[J]. Jpn J Infect Dis, 2017, 70(4): 378-382. DOI:10.7883/yoken.JJID.2016.234 |
| [5] |
Li YH, Luo Q, Shi XL, et al. Phenotypic and genotypic chara- cterization of clinical enterotoxigenic Escherichia coli isolates from Shenzhen, China[J]. Foodborne Pathog Dis, 2017, 14(6): 333-340. DOI:10.1089/fpd.2016.2233 |
| [6] |
García-Meniño I, García V, Mora A, et al. Swine enteric colibacillosis in Spain: pathogenic potential of mcr-1 ST10 and ST131 E. coli isolates[J]. Front Microbiol, 2018, 9: 2659. DOI:10.3389/fmicb.2018.02659 |
| [7] |
Huang Z, Pan HJ, Zhang PP, et al. Prevalence and antimicrobial resistance patterns of diarrheagenic Escherichia coli in Shanghai, China[J]. Pediatr Infect Dis J, 2016, 35(8): 835-839. DOI:10.1097/INF.0000000000001190 |
| [8] |
李春, 俞俊岭. 安徽地区腹泻患者感染肠致病大肠埃希氏菌的耐药特征[J]. 中国人兽共患病学报, 2019, 35(8): 732-737. Li C, Yu JL. Resistance characteristics of enteropathogenic Escherichia coli isolated from feces of diarrhea patients in Anhui, China[J]. Chinese Journal of Zoonoses, 2019, 35(8): 732-737. |
| [9] |
白向宁, 赵爱兰, 夏胜利, 等. 非O157产志贺毒素大肠杆菌分离株的多位点序列分型研究[J]. 中国人兽共患病学报, 2012, 28(6): 544-548, 560. Bai XN, Zhao AL, Xia SL, et al. Multilocus sequence typing of non-O157 Shiga toxin-producing Escherichia coli isolates[J]. Chinese Journal of Zoonoses, 2012, 28(6): 544-548, 560. |
| [10] |
Chen Y, Chen X, Zheng S, et al. Serotypes, genotypes and antimicrobial resistance patterns of human diarrhoeagenic Escherichia coli isolates circulating in southeastern China[J]. Clin Microbiol Infect, 2014, 20(1): 52-58. DOI:10.1111/1469-0691.12188 |
| [11] |
张子科, 赖圣杰, 余建兴, 等. 我国2012-2015年门诊腹泻患者中致泻性大肠埃希菌流行特征分析[J]. 中华流行病学杂志, 2017, 38(4): 419-423. Zhang ZK, Lai SJ, Yu JX, et al. Epidemiological characteristics of diarrheagenic Escherichia coli among diarrhea outpatients in China, 2012-2015[J]. Chinese Journal of Epidemio-logy, 2017, 38(4): 419-423. DOI:10.3760/cma.j.issn.0254-6450.2017.04.002 |
| [12] |
李春, 陈国平, 孟昭倩, 等. 2015-2019年安徽省致泻性大肠埃希菌分型及耐药性分析[J]. 中华疾病控制杂志, 2020, 24(10): 1154-1159. Li C, Chen GP, Meng ZQ, et al. Typing and antimicrobial resistance analysis of diarrheogenic Escherichia coli in Anhui Province from 2015 to 2019[J]. Chinese Journal of Disease Control & Prevention, 2020, 24(10): 1154-1159. |
| [13] |
石挺丽, 黄建华, 李秀芬, 等. 2014-2015年广州地区腹泻患儿和健康儿童致泻性大肠埃希菌流行特征及耐药分析[J]. 中华疾病控制杂志, 2016, 20(4): 329-332. Shi TL, Huang JH, Li XF, et al. Epidemiological characteri-zation and antimicrobial resistance of diarrhea Escherichia coli from acute diarrheal and healthy children in Guangzhou City, 2014-2015[J]. Chinese Journal of Disease Control & Prevention, 2016, 20(4): 329-332. |
| [14] |
杨杰, 李文, 冀国强, 等. 顺义区2018年腹泻病例粪便中致泻大肠埃希氏菌耐药监测结果分析[J]. 安徽预防医学杂志, 2020, 26(5): 386-389. Yang J, Li W, Ji GQ, et al. Analysis of drug resisitance of diarrheal Escherichia coli in stool of diarrhea cases in Shunyi Diarrheal in 2018[J]. Anhui Journal of Preventive Medicine, 2020, 26(5): 386-389. |
| [15] |
韩仁如, 郑永贵, 杨洋, 等. 2019年复旦大学附属华山医院细菌耐药性监测[J]. 中国感染与化疗杂志, 2020, 20(5): 457-464. Han RR, Zheng YG, Yang Y, et al. Surveillance of antimicrobial resistance among bacterial strains isolated from Huashan Hospital, Fudan University in 2019[J]. Chinese Journal of Infection and Chemotherapy, 2020, 20(5): 457-464. |
| [16] |
Okeke IN, Wallace-Gadsden F, Simons HR, et al. Multi-locus sequence typing of enteroaggregative Escherichia coli isolates from Nigerian children uncovers multiple lineages[J]. PLoS One, 2010, 5(11): e14093. DOI:10.1371/journal.pone.0014093 |



